Modelling my brain
March 2021
This is a 3d model I made of my own brain, and the story of how I made it.
In early 2013 I took part in a medical experiment to explore the effects of hypoxia (lack of oxygen) on the brain. For 22 hours I lived in a plastic tent in a hospital whilst breathing 12% oxygen, equivalent to around 4000m altitude. During this time I also spent a total of four and half hours in an MRI machine, breathing the oxygen-deficient air through a face mask.


As a mountaineer, I'd typically ascend to this altitude over a period of days to allow my body to acclimatise. For this study we reached "4000m" in under 60 seconds, which wasn't pleasant. It was a night of strong headaches and little sleep, followed by a good dose of vomiting for my study partner Jamie. In fact of the 12 victims participants, 4 withdrew early due to onset of acute mountain sickness.
Generating reduced O₂ levels was achieved using oxygen hypoxicators. Typically used by athletes for altitude training, they're basically just the exhaust pipe of the far more common oxygen concentrator. The system consisted of two of these in series along with a Bair Hugger to force positive pressure airflow into the tent to compensate for the lack of airtight seal. The experimental setup was very Apollo-13, right down to the jug of soda lime chips to absorb carbon-dioxide, connected with a plastic bag and duct-tape due to a mismatch in pipe diameters.
Luckily, the setup worked flawlessly.


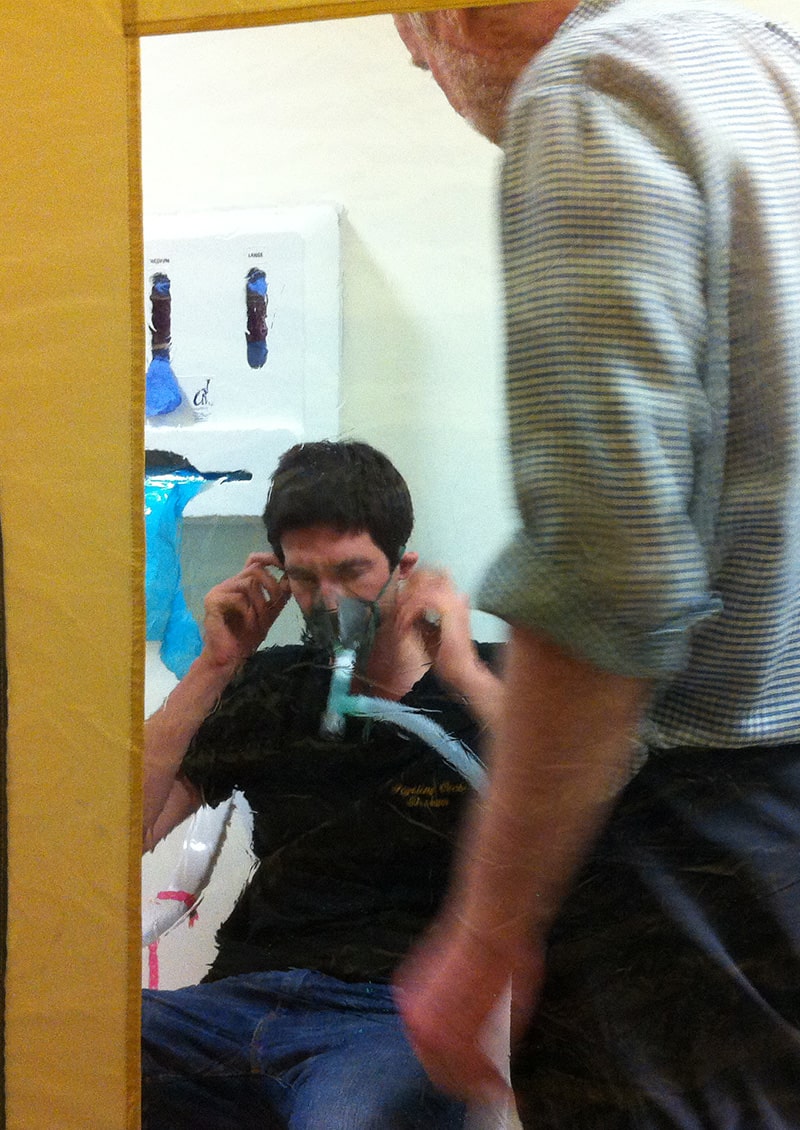
I've no doubt that the research resulted in some useful contribution to science. But far more interesting for me was the CD of 7000 slices of my brain I walked away with. 8 years later, it's time to put them to good use!
Slicing the brain
The study explored how blood flow to the brain changes during the development of HACE (High Altitude Cerebral Edema). HACE at altitude can easily be fatal and is treated using dexamethasone, which you might recognise from its use in treating COVID-19. During the study I underwent 6x 45 minute MRI scans, which generated a lot of data including some fascinating blood vessel scans:
The data came in the form of a DICOM directory, most of which isn't easily navigable in Windows so I used the free DICOM viewer Invesalius to load in the datasets. A typical full-head scan contained around 200 slices in each of the three axes. Invesalius will interpret and stich the slices into a 3d model, but the resultant model includes everything: skin, eyeballs, brain matter, blood vessels, muscle...
Distinguishing between these types of tissue can be achieved by masking the slices. Invesalius came with a few presets (e.g. Compact bone, Enamel, Fat Tissue) but there sadly wasn't a one-click "brain" option. The presets appear to generate a mask based on a portion of the image greyscale, which must equate to the tissue density from the MRI scan. Invesalius includes some quite advanced segmentation tools for creating mask watersheds and region growing as well as set of wild ray-tracing visualisation tools for quick visualisation of the dataset.
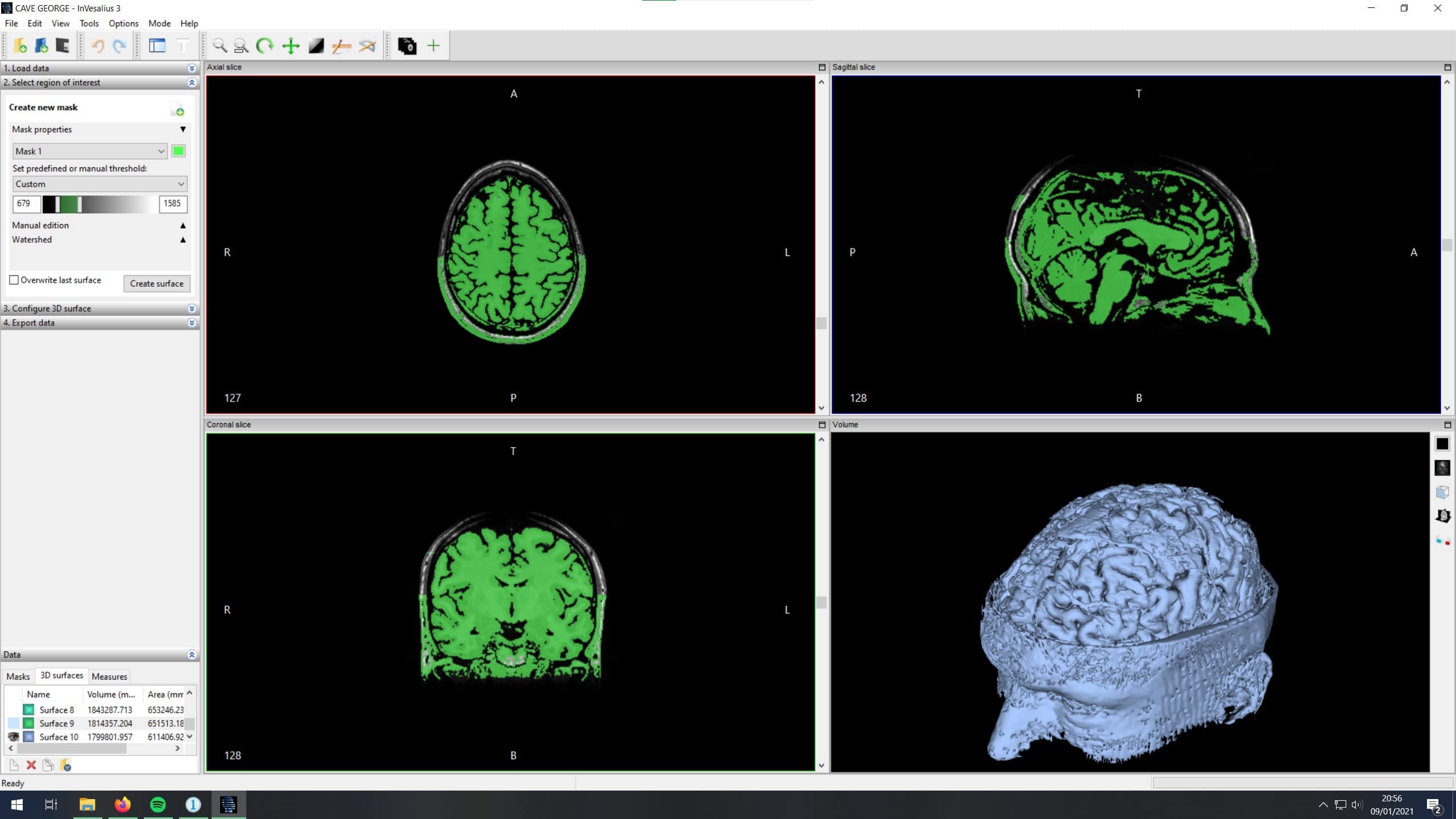
Despite the excellent documentation, in the end I never got to grips with the automated tools.
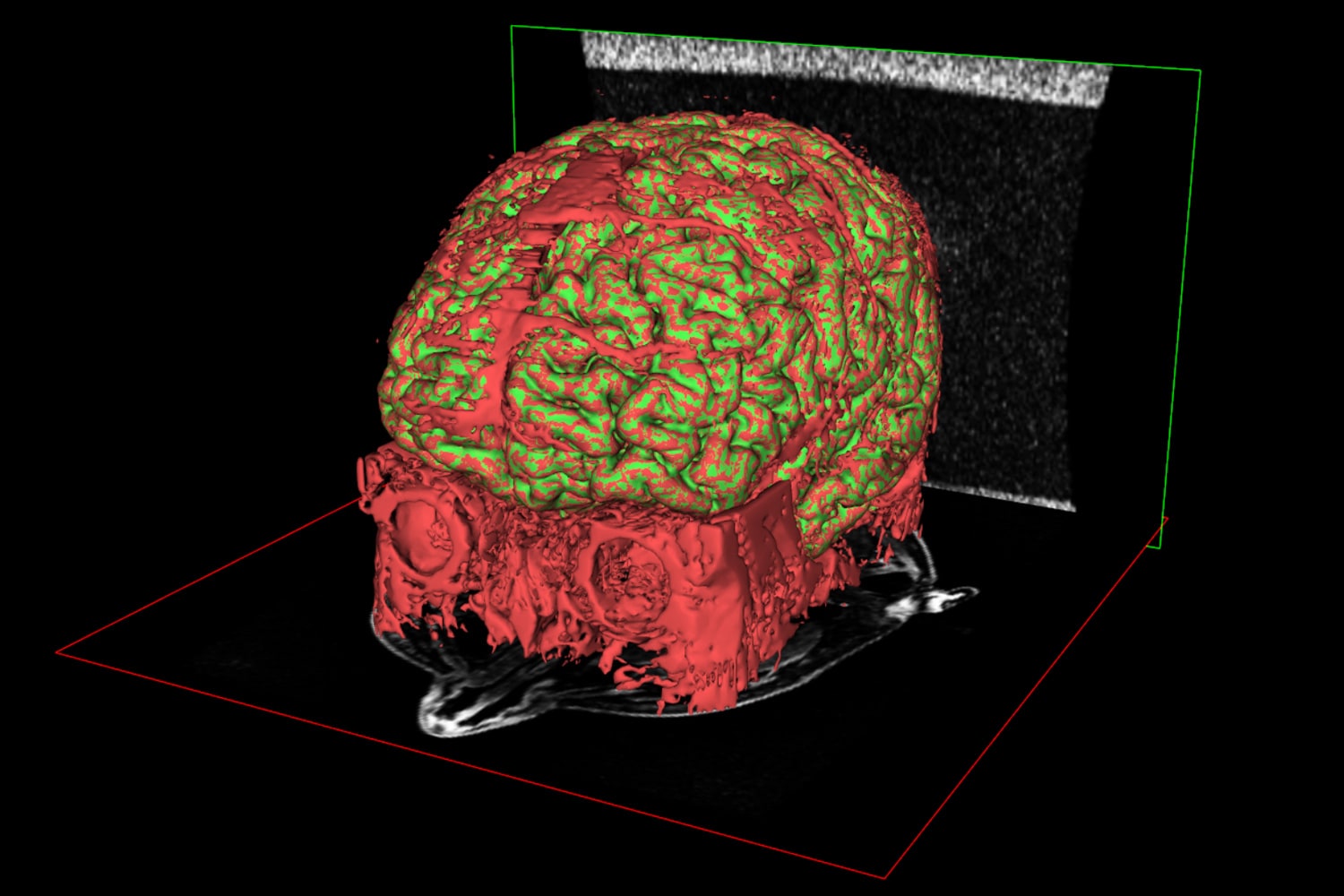
Instead, it took around 30 hours to manually mask the brain tissue from all 600 images. The process required regular cross-referencing between the 2d slices and 3d model to ensure I didn't perform an accidental lobotomy. Using your own brain to examine your own brain can really mess with your head.
Invesalius exports clean STL surfaces, but working on the model as a sequence of 2D slices left a number of small blood vessels clinging to the surface, particular those which weren't aligned to one of the principle axes. A couple of hours of clean up in Meshmixer was enough to prepare the final data.


Building the brain
The great thing about medical data is that it is inherently 1:1, which makes printing to scale trivial. The model came out significantly heavier than anticipated because whilst the gyri have a light infill, the model is fully detailed with gyri and sulci all the way through. Thanks to my friend Michael Domhardt for donating 70 hours of printing time on his Prusa Bear i3.
And now I have no idea what to do with it.




